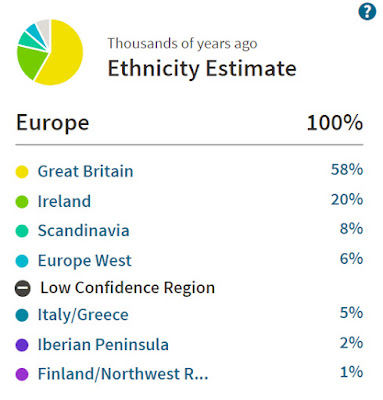
I wrote back in June about my updated Ethnicity Estimate at AncestryDNA. Yesterday AncestryDNA rolled out the updates to everyone in their database. Many people will find that the changes are quite dramatic. I went from being just 21% Great Britain to 94% England and Wales, and my results are now a much better reflection of my recent ancestry within the last few hundred years. There have been a few tweaks since I got my results and the England and Wales cluster has now been renamed as England, Wales and Northwestern Europe.
The improvements have been made possible by the inclusion of many more people in the reference panel, which has now gone up from 3,000 to 16,000 samples. Previously Ancestry had just 111 samples from Great Britain, 138 from Ireland and 166 from Europe West. Now they have 1,519 samples from England, Wales and Northwestern Europe, 500 from Ireland and Scotland, 1,407 from France and 2072 from Germanic Europe. AncestryDNA are also using a different methodology and are comparing long stretches of linked markers rather than single markers in isolation. This means that the results are a reflection of our more recent ancestry within the last 500 to 1000 years rather than our distant ancestry from one thousand or more years ago.
AncestryDNA have written a White Paper explaining the methodology, which includes details of all the reference populations used. They will also be publishing a scientific paper about their methods.
Most people with British and Irish ancestry have found that their results are greatly improved and are much more in line with their known ancestry. The results will be more mixed for people from other countries. You can only be matched to the populations in the reference panel so if your country is not represented you will be matched to the next closest population. For example, AncestryDNA now has reference populations for Norway, Sweden and Finland but no distinct dataset for Denmark. Danes are therefore likely to get matched with Norway and Sweden or England, Wales and Northwestern Europe.
India, with a vast and diverse population of over 1.3 billion people, is poorly represented with just 65 samples from Western and Central India. There is also still a long way to go to get more meaningful results for people with African ancestry. There is more genetic diversity in Africa than in the rest of the world combined, which means that much larger reference panels are needed to capture this diversity. Ancestry are addressing this problem by starting an African Diversity Project, and we can look forward to further improvements in the years to come.
I always used to say that "ethnicity" estimates should be taken with a large pinch of salt and are really only of entertainment value, but we are now starting to get the stage where the results for some people can provide a reasonable approximation of their ancestry. If you've already done your family history research, the results won't tell you anything more than you already know, but at least there should now be a lot less confusion. As more populations are added to the reference panels we can expect to see similar improvements for other populations.
Update 15th September 2018
AncestryDNA will be presenting a poster at the ASHG conference in San Diego in October on Polly, the algorithm they are using for their updated ethnicity estimates. Here are the details:
The ASHG abstracts can be searched here.
Further reading
I've provided links below to the various official documents from AncestryDNA along with links to a few other blogs which might be of interest.
The improvements have been made possible by the inclusion of many more people in the reference panel, which has now gone up from 3,000 to 16,000 samples. Previously Ancestry had just 111 samples from Great Britain, 138 from Ireland and 166 from Europe West. Now they have 1,519 samples from England, Wales and Northwestern Europe, 500 from Ireland and Scotland, 1,407 from France and 2072 from Germanic Europe. AncestryDNA are also using a different methodology and are comparing long stretches of linked markers rather than single markers in isolation. This means that the results are a reflection of our more recent ancestry within the last 500 to 1000 years rather than our distant ancestry from one thousand or more years ago.
AncestryDNA have written a White Paper explaining the methodology, which includes details of all the reference populations used. They will also be publishing a scientific paper about their methods.
Most people with British and Irish ancestry have found that their results are greatly improved and are much more in line with their known ancestry. The results will be more mixed for people from other countries. You can only be matched to the populations in the reference panel so if your country is not represented you will be matched to the next closest population. For example, AncestryDNA now has reference populations for Norway, Sweden and Finland but no distinct dataset for Denmark. Danes are therefore likely to get matched with Norway and Sweden or England, Wales and Northwestern Europe.
India, with a vast and diverse population of over 1.3 billion people, is poorly represented with just 65 samples from Western and Central India. There is also still a long way to go to get more meaningful results for people with African ancestry. There is more genetic diversity in Africa than in the rest of the world combined, which means that much larger reference panels are needed to capture this diversity. Ancestry are addressing this problem by starting an African Diversity Project, and we can look forward to further improvements in the years to come.
I always used to say that "ethnicity" estimates should be taken with a large pinch of salt and are really only of entertainment value, but we are now starting to get the stage where the results for some people can provide a reasonable approximation of their ancestry. If you've already done your family history research, the results won't tell you anything more than you already know, but at least there should now be a lot less confusion. As more populations are added to the reference panels we can expect to see similar improvements for other populations.
Update 15th September 2018
AncestryDNA will be presenting a poster at the ASHG conference in San Diego in October on Polly, the algorithm they are using for their updated ethnicity estimates. Here are the details:
PgmNr 2772/W: High-throughput local ancestry inference reveals fine-scale population history
Authors:A. Sedghifar1; S. Song1; Y. Wang1; K. Noto1; J. Byrnes1; E.L. Hong1; K.G. Chahine1; C.A. Ball2
Affiliations:
1) AncestryDNA, San Francisco, CA.; 2) AncestryDNA, Lehi, UT.
An individual’s genome can be viewed as a mosaic of haplotype blocks from different ancestral origins, the sizes of which depend on the timing of admixture events. Recovering the length of these local ancestry blocks, together with their ethnic origin, provides information on the admixture and recombination events that shape current day genomes, thus shedding light on personal history as well as population history. As genomic databases rapidly approach sizes on the order of millions of genomes, there is an increased demand for super efficient approaches to identifying local ancestry blocks. Our team has developed Polly, an ultra fast algorithm for estimating genome-wide ancestry proportions in admixed individuals. Here, we present a modification of the Polly algorithm for accurately inferring local ancestry blocks. We evaluated the performance of our algorithm on simulated admixed individuals, and also assessed accuracy of estimated tract length distributions in admixed populations. Finally, we applied our method to estimate tract length distributions in historically admixed African American and Latin American populations.The poster can be seen here.
The ASHG abstracts can be searched here.
Further reading
I've provided links below to the various official documents from AncestryDNA along with links to a few other blogs which might be of interest.
AncestryDNA links
- Major enhancement to AncestryDNA's Ethnicity Estimates by Leah Larkin, The DNA Geek
- Finally the AncestryDNA update we've all been waiting for by Donna Rutherford
- AncestryDNA revises ethnicity estimates by Blaine Bettinger, The Genetic Genealogist
- AncestryDNA ethnicity estimates updated by Judy Russell, The Legal Genealogist
- Finally, Ancestry's new ethnicity estimates by Kitty Cooper