Black Friday is a concept that's been imported from America. It's the day after the US Thanksgiving holiday and traditionally the day when Americans start their Christmas shopping. The retailers often provide special deals to lure everyone to the shops. Lots of shops and websites are now trying to do the same in the UK and, whether we like or not, Black Friday is here to stay. For genetic genealogists benefit of this gimmick is that the DNA testing companies are jumping onto the bandwagon and offering DNA kits at reduced prices not just for their US customers but for people in the UK, Ireland, Australia, New Zealand and other countries too.
I was going to do a blog post summarising all the current Black Friday offers but Michelle Leonard has already beaten me to it. If you want to find out about all the special deals make sure you check out Michelle's blog post Black Friday DNA Sales 2017. She will update the page with new offers as they become available. Note that many of these discounts are for a very limited period only and will expire on Monday which is now known as Cyber Monday – the day when many people (though not me!) start their online Christmas shopping.
For full details of all the Family Tree DNA sale prices, including all the discounted upgrades, see my previous post FTDNA's 13th International Genetic Genealogy Conference and the FTDNA sale.
If you've not yet had your DNA tested or want to test additional family members or you want to get your DNA into a different database now is a great time to test.
The day-to-day activities of the Cruwys/Cruse one-name study with occasional diversions into other topics of interest such as DNA testing and personal genomics
Thursday, 23 November 2017
Monday, 13 November 2017
FTDNA's 13th International Genetic Genealogy Conference and the FTDNA sale
At the weekend Family Tree DNA held their 13th International Genetic Genealogy Conference for group administrators. You can see the full programme here. I've been following the news from the conference on Twitter by checking out the hashtag #FTDNA2017. Thank you to Katherine Borges, Lisa Janine Cloud, Louis Kessler and Marilyn Souders for all the tweets and photos.
Some of the conference attendees have blogged about the conference. I've provided a list below which I will update if further articles become available.
Conference reports from Jennifer Zinck
Jennifer always writes wonderfully detailed notes from the conference and I recommend reading her articles in their entirety. It's almost like being there in person!
- 13th International Genetic Genealogy Conference – Saturday
- 13th International Genetic Genealogy Conference – Sunday
Louis Kessler is a first-time conference attendee. He provides an interesting perspective and has also shared a selection of photographs.
At the close of the conference FTDNA announced a sale which will run until the end of the year. Upgrades are also included in the sale. Here is a chart showing the sale prices.
Note that, in addition to the above prices, FTDNA charges $12.95 for shipping. Kits for customers in the US are sent out by DHL and the cost includes return shipping. Customers in other countries will need to pay the return postage separately.
There is also a 15% discount on SNPs and SNP packs for the sale period.
FTDNA have given existing customers discount codes (coupons) which can be used to get a further reduction on the sale price. You'll need to log into your FTDNA account and click on the Holiday Reward to see your offer. As always, a number of people have taken the initiative to set up collaborative spreadsheets where unused coupons can be shared and exchanged.
A spreadsheet is available from this link that was shared on the Haplogroup Facebook page.
There are also other spreadsheets being maintained by specific groups such as the U106 group on Yahoo.
- FTDNA’s 13th International Genetic Genealogy Conf, Day 1
- FTDNA’s 13th International Genetic Genealogy Conf Day 2
- FTDNA’s 13th International Genetic Genealogy Conf Day 3
- FTDNA's 13th IGG conference lab tour
Conference report from Judy Russell
- What's not in our genes An overview of Michael Hammer's talk
Conference report from Rob van Drie
Presentations
The slides from some of the presentations will be made available to project administrators.
Maurice Gleeson has shared his two conference presentations on his YouTube channel:
FTDNA saleAt the close of the conference FTDNA announced a sale which will run until the end of the year. Upgrades are also included in the sale. Here is a chart showing the sale prices.
| Individual Tests | Sale price | Regular price |
Family Finder
(FF)
|
$59 | $89 |
mtFull Sequence (FMS)
|
$169 | $199 |
Y-37
|
$129 | $169 |
Y-67
|
$229 | $268 |
Y-111
|
$299 | $359 |
Test Bundles
|
Sale price | Regular price |
Family Finder + Y-37
|
$178 | $238 |
Family Finder + Y-67
|
$278 | $337 |
FF + mtFull Sequence
|
$218 | $268 |
FF + Y-67 + mtFull Sequence
|
$442 | $536 |
Upgrades
|
Sale price | Regular price |
mt/mtPlus to
FMS
|
$119 | $159 |
Big
Y
|
$475 | $575 |
Y-12 to Y-37
|
$69 | $109 |
Y-25 to Y-37
|
$35 | $59 |
Y-37 to Y-67
|
$79 | $109 |
Y-37 to
Y-111
|
$168 | $228 |
Y-67 to
Y-111
|
$99 | $129 |
Note that, in addition to the above prices, FTDNA charges $12.95 for shipping. Kits for customers in the US are sent out by DHL and the cost includes return shipping. Customers in other countries will need to pay the return postage separately.
There is also a 15% discount on SNPs and SNP packs for the sale period.
FTDNA have given existing customers discount codes (coupons) which can be used to get a further reduction on the sale price. You'll need to log into your FTDNA account and click on the Holiday Reward to see your offer. As always, a number of people have taken the initiative to set up collaborative spreadsheets where unused coupons can be shared and exchanged.
A spreadsheet is available from this link that was shared on the Haplogroup Facebook page.
There are also other spreadsheets being maintained by specific groups such as the U106 group on Yahoo.
Wednesday, 8 November 2017
23andMe passes three million milestone
I've not been able to find any official announcement but 23andMe now state on the About Us page on their website that they have genotyped over three million customers. This figure is also cited in an article on genealogy tourism published in USA Today on 3rd November 2017.
The 23andMe database stood at two million in April 2017 when it was announced that they had received FDA approval to provide health reports for their US customers. That means they have tested one million people in the last seven months.
It was announced in October that 23andMe had "launched an advertising blitz to dramatically expand its customer base to 10 million people". How long will it be before they reach that goal?
Thanks to Ritchie Hansen for alerting me to the USA Today story in the Genetic Genealogy Tips and Techniques group on Facebook.
The 23andMe database stood at two million in April 2017 when it was announced that they had received FDA approval to provide health reports for their US customers. That means they have tested one million people in the last seven months.
It was announced in October that 23andMe had "launched an advertising blitz to dramatically expand its customer base to 10 million people". How long will it be before they reach that goal?
Thanks to Ritchie Hansen for alerting me to the USA Today story in the Genetic Genealogy Tips and Techniques group on Facebook.
Thursday, 2 November 2017
AncestryDNA updates – six million customers, a new DNA Story layout and changes to the consent process
There have been a few changes at Ancestry DNA in the last few days. They've started to roll out a new presentation for their "ethnicity" reports and Genetic Communities. The two features are now integrated which makes them much easier to use. This is what my new home page looks like. The admixture percentages have not changed but I am encouraged to click through to view my DNA Story.
Here is my new DNA Story page. By default the low-confidence regions are not shown, but I've expanded them in this screenshot.
I currently have one Genetic Community for Southern England. That is now nested inside the Great Britain region.
The admixture component that was previously labelled Ireland has now been renamed as Ireland/Scotland/Wales. However, this component actually covers much of England as well, and overlaps with the Southern England Genetic Community, as can be seen below.
The timeline is now conveniently located at the bottom of the page. It's easy to click through and read the historical detail for the different periods. Here's my timeline from 1850 which shows the migrations of the people in my Southern England community. This is the era of peak emigration, particularly to Australia, New Zealand, Canada and America, and it's fascinating to see how these connections are showing up in the DNA.
Some people are also seeing Migrations listed below their "ethnicity" report. This only applies if the person's genetic community is not part of their admixture regions.
AncestryDNA database now at six million and changes to the consent process
AncestryDNA have also announced that their database has reached six million. They had five million people in their database at the beginning of August 2017, which means they've added one million to their database in less than three months. The good news is that much of that growth has taken place in the UK, Ireland, Australia and New Zealand.
At the same time AncestryDNA have also made some changes to the DNA cousin-matching service, as described in this blog post. Most importantly, customers now have the ability to opt out of cousin matching, and new customers have to actively consent to participate in the relative-matching database. This is what the test settings now look like for existing customers.
This is the message you get if you want to change the settings for your DNA matches.
Here is the page in the activation process for new testers where you have to consent to "see and be seen" by your DNA matches.
This is what the home page now looks like if the tester decides not to opt in to matching. (Many thanks to Michelle Leonard for these two screenshots.)
This is a very welcome change, and I'm just surprised that it didn't happen sooner. This brings AncestryDNA into line with 23andMe and Family Tree DNA, both of whom require the tester to opt in to be included in the matching database.
I always warn my fellow genealogists to be prepared for the unexpected. With the growth in the DNA databases we are finding many examples of surprise DNA matches. Some people are getting unexpected matches with previously unknown close relatives, and sometimes even with unknown siblings and parents. Much of the focus of the AncestryDNA marketing campaigns has been on the admixture percentages, and this has helped to bring in a whole new demographic to DNA testing, many of whom have then been inspired to start researching their family tree. However, many of these people don't realise that the test also has a direct genealogical application and can be used for cousin matching. I've personally come across a couple of instances of people testing to get the admixture percentages and then finding by chance that they had an unknown parent or child in the database. These discoveries were made in the most inappropriate circumstances and the embarrassment could have been avoided if there had been a proper consent process in place at the time. It's important that everyone who participates in the DNA matching database understands what they are doing and that they are forewarned about the implications so that they can make their own choices based on their individual circumstances. Not everyone is willing or ready to discover unknown relatives and their wishes should be respected.
Further reading
Here is my new DNA Story page. By default the low-confidence regions are not shown, but I've expanded them in this screenshot.
I currently have one Genetic Community for Southern England. That is now nested inside the Great Britain region.
The admixture component that was previously labelled Ireland has now been renamed as Ireland/Scotland/Wales. However, this component actually covers much of England as well, and overlaps with the Southern England Genetic Community, as can be seen below.
The timeline is now conveniently located at the bottom of the page. It's easy to click through and read the historical detail for the different periods. Here's my timeline from 1850 which shows the migrations of the people in my Southern England community. This is the era of peak emigration, particularly to Australia, New Zealand, Canada and America, and it's fascinating to see how these connections are showing up in the DNA.
Some people are also seeing Migrations listed below their "ethnicity" report. This only applies if the person's genetic community is not part of their admixture regions.
AncestryDNA database now at six million and changes to the consent process
AncestryDNA have also announced that their database has reached six million. They had five million people in their database at the beginning of August 2017, which means they've added one million to their database in less than three months. The good news is that much of that growth has taken place in the UK, Ireland, Australia and New Zealand.
At the same time AncestryDNA have also made some changes to the DNA cousin-matching service, as described in this blog post. Most importantly, customers now have the ability to opt out of cousin matching, and new customers have to actively consent to participate in the relative-matching database. This is what the test settings now look like for existing customers.
This is the message you get if you want to change the settings for your DNA matches.
Here is the page in the activation process for new testers where you have to consent to "see and be seen" by your DNA matches.
I always warn my fellow genealogists to be prepared for the unexpected. With the growth in the DNA databases we are finding many examples of surprise DNA matches. Some people are getting unexpected matches with previously unknown close relatives, and sometimes even with unknown siblings and parents. Much of the focus of the AncestryDNA marketing campaigns has been on the admixture percentages, and this has helped to bring in a whole new demographic to DNA testing, many of whom have then been inspired to start researching their family tree. However, many of these people don't realise that the test also has a direct genealogical application and can be used for cousin matching. I've personally come across a couple of instances of people testing to get the admixture percentages and then finding by chance that they had an unknown parent or child in the database. These discoveries were made in the most inappropriate circumstances and the embarrassment could have been avoided if there had been a proper consent process in place at the time. It's important that everyone who participates in the DNA matching database understands what they are doing and that they are forewarned about the implications so that they can make their own choices based on their individual circumstances. Not everyone is willing or ready to discover unknown relatives and their wishes should be respected.
Further reading
- Your DNA has a story to tell - the official blog post from the Ancestry team
- Donna Rutherford has written a nice blog post about her new DNA Story feature at AncestryDNA.
Friday, 27 October 2017
23andMe now offers an ancestry-only DNA test in Europe and Canada
23andMe offer a very interesting test which includes both health and ancestry reports. However, because of the high cost of the test, it has never been practical to use it in the UK for genealogy purposes. Our American friends have had the option to buy an ancestry-only test for $99 for a while now but that facility has not been available in other countries. That situation has all changed some time in the last few days though we don't know exactly when this change was rolled out. In the countries where 23andMe offer health reports it is now possible to order an ancestry-only test at a lower and affordable price. Here are the new prices.
For the remaining countries where the 23andMe test is sold, an international ancestry-only test is available for US $99 plus shipping. The cost of the international test has also been reduced in recent months. Previously this test cost $149 outside the US. Shipping costs vary from country to country, but these costs have also been greatly reduced. You can find full details of international availability and shipping costs by country on the 23andMe page in the ISOGG Wiki.
These reduced prices are really good news for genetic genealogists. At £79 the 23andMe test represents very good value for money. In addition to receiving cousin matches and a biogeographical ancestry analysis, the test will also give you haplogroup assignments (Y-DNA and mtDNA for males; mtDNA only for females). The Y-DNA and mtDNA reports can't be used for matching purposes but they are very useful if you just want a general overview of your ancestry. 23andMe are also the only genetic genealogy company that provides reports on your Neanderthal ancestry.
Here is a screenshot of my 23andMe ancestry reports.
This is an extract from my Neanderthal ancestry report. There is a lot more detail in these reports which there is not space to include here.
The Neanderthal report also includes this chromosome map showing the location of the Neanderthal variants in your genome.
It was reported recently that 23andMe are going to embark on an advertising blitz to "dramatically expand" their customer base to 10 million people. Let's hope that the launch of the ancestry-only test will encourage many new people to test with 23andMe, and help to expand the customer database outside the US.
Thanks to Michelle Leonard for alerting me to the introduction of 23andMe's new ancestry-only tests. You can read Michelle's blog post on the subject here.
| Country | Ancestry | Ancestry and health | Return shipping |
| UK | £79 | £149 | £9.99 |
| Denmark, Finland, Ireland, Sweden and the Netherlands | €99 | €169 | €9.99 |
| Canada | C$129 | C$249 | C$19.95 |
| US | $99 | $199 | $9.95 |
For the remaining countries where the 23andMe test is sold, an international ancestry-only test is available for US $99 plus shipping. The cost of the international test has also been reduced in recent months. Previously this test cost $149 outside the US. Shipping costs vary from country to country, but these costs have also been greatly reduced. You can find full details of international availability and shipping costs by country on the 23andMe page in the ISOGG Wiki.
These reduced prices are really good news for genetic genealogists. At £79 the 23andMe test represents very good value for money. In addition to receiving cousin matches and a biogeographical ancestry analysis, the test will also give you haplogroup assignments (Y-DNA and mtDNA for males; mtDNA only for females). The Y-DNA and mtDNA reports can't be used for matching purposes but they are very useful if you just want a general overview of your ancestry. 23andMe are also the only genetic genealogy company that provides reports on your Neanderthal ancestry.
Here is a screenshot of my 23andMe ancestry reports.
This is an extract from my Neanderthal ancestry report. There is a lot more detail in these reports which there is not space to include here.
The Neanderthal report also includes this chromosome map showing the location of the Neanderthal variants in your genome.
It was reported recently that 23andMe are going to embark on an advertising blitz to "dramatically expand" their customer base to 10 million people. Let's hope that the launch of the ancestry-only test will encourage many new people to test with 23andMe, and help to expand the customer database outside the US.
Thursday, 26 October 2017
Living DNA updates - free transfers and the launch of the One Family One World project
Living DNA have just launched their new One Family One World project and are now accepting transfers from people who have tested with other companies. Here is the text of the e-mail I received from Living DNA:
Here at Living DNA, we have been working on an exciting new project that we have officially launched today. The One Family One World project. As you've been a great support to Living DNA I wanted to let you know first.
Our One Family One World project is the first of its kind to attempt to analyse people’s DNA results from around the world allowing them to see where they fit into a One World Family Tree, demonstrating how everyone is related if you go back far enough in time and produce an in-country regional breakdown of DNA from around the world.
The project involves the use of proprietary technology, artificial intelligence and machine learning, and will see tens of thousands of computers working together to identify distinctive and shared patterns in people’s DNA.
Eventually we will be running in-country regional projects across the world, today people can join no matter where they are from and we already have a number of specific projects that can be seen here - https://www.livingdna.com/one-family/research.
You can find out a lot more information on the project, and how to get involved by visiting the website at www.livingdna.com/onefamily. If you know anyone that’s already taken a DNA test they can upload their DNA for Free and will benefit from a new type of DNA Matching in mid 2018.
Your support and opinion matters to us, so we would love to hear your feedback on the project. Also, let us know if you would like to get involved in any way - we always value any way in which people would like to work with us.
We’ve put together an introduction blog post that you can read https://www.livingdna.com/en-gb/blog/291/living-dna-demonstrating-how-we-re-all-connected-through-one-world-family-tree
We hope you’re as excited as we are on this new 5-year project, and I look forward to hearing from you all soon.
Warm regards,
David Nicholson & Hannah Mordern
ps. Apologies for the group email and do feel free to circulate this to other people
pps. DNA Matching is in final stages of testing for Living DNA clients, we are still aiming to get this live by the end of the year
ppps. The press release is also online https://www.livingdna.com/en-gb/press-releases/290/dna-firm-aims-build-one-world-family-tree
pppps. People taking part and uploading their DNA for free don't get the regional UK breakdown just DNA matching when live and all the other things that will come out of the project. Therefore its not a substitute for a Living DNA test.
You can watch a video about the One Family project here.
If you are reading this by e-mail you can click here instead.
Living DNA already have projects for Ireland and Germany where they are collecting samples from people with four grandparents from the same region so it's good to see that this concept has been extended to other countries. With better sampling and larger reference datasets we can expect to see continuing improvements to the biogeographical ancestry reports with the promise of meaningful breakdowns at the regional and subregional levels.
Living DNA were the sponsors of the evening meal for the ISOGG Big Day Out in Dublin at Genetic Genealogy Ireland. I had the chance to talk to Martin Blythe of Living DNA about their forthcoming matching service. They will be delivering phased matching but they are also adopting an exciting and innovative new approach. It will be very interesting to see how their matching works out and how it compares with the services provided by the other testing companies.
If you don't want to do the transfer and prefer to get tested on Living DNA's new Orion chip (a customised version of Illumina's Global Screening Array chip) then the test is currently on sale at the reduced price of £99 ($99 in the US, €129,00 in the Eurozone). Check out the Living DNA website for prices in other countries. If you have four grandparents born within 50 miles of one another you will be eligible for a special discount on a new test.
Update 31 October 2017
Living DNA have published the following notice in the Living DNA Users' Group on Facebook in response to questions received about the One Family One World Project:
If you are reading this by e-mail you can click here instead.
Living DNA already have projects for Ireland and Germany where they are collecting samples from people with four grandparents from the same region so it's good to see that this concept has been extended to other countries. With better sampling and larger reference datasets we can expect to see continuing improvements to the biogeographical ancestry reports with the promise of meaningful breakdowns at the regional and subregional levels.
Living DNA were the sponsors of the evening meal for the ISOGG Big Day Out in Dublin at Genetic Genealogy Ireland. I had the chance to talk to Martin Blythe of Living DNA about their forthcoming matching service. They will be delivering phased matching but they are also adopting an exciting and innovative new approach. It will be very interesting to see how their matching works out and how it compares with the services provided by the other testing companies.
If you don't want to do the transfer and prefer to get tested on Living DNA's new Orion chip (a customised version of Illumina's Global Screening Array chip) then the test is currently on sale at the reduced price of £99 ($99 in the US, €129,00 in the Eurozone). Check out the Living DNA website for prices in other countries. If you have four grandparents born within 50 miles of one another you will be eligible for a special discount on a new test.
Update 31 October 2017
Living DNA have published the following notice in the Living DNA Users' Group on Facebook in response to questions received about the One Family One World Project:
Hi Everyone,
The One Family One World, five year scientific research project has started with great success with thousands of people delighted to be supporting the project and its purpose.
As with all scientific projects results will take time as it requires the combined analysis of hundreds of thousands of DNA samples.
We’ve received some questions that we want to clarify.
How long will the project take?
This is a five year research project and results will be announced in phases, the exact timing of the results will depend on how many people from different parts of the world take part. A critical mass for each area is required and for the overall One Family Tree.
A number of people who requested to join the project and uploaded their data received an incorrect estimated completion date. These have now been updated to reflect when global DNA matching will be available, which is by summer 2018. This is not the date the project will be complete.
What do I get from taking part?
The purpose of the project is to help scientific research, and ultimately build a global family tree. To say thank you for your support, each person will be able to choose to see how they match and connect to other people both within the project and with Living DNA clients. What we can’t offer for free is your ethnic breakdown simply due to the significant data analysis costs.
But if you buy a Living DNA kit, then you can take part in the project, receive matching when it is live and see your mtDNA/YDNA and ethnicity breakdowns.
How do I get my ethnicity breakdown?
Living DNA are working on delivering this, so that people who’ve already taken a DNA test can upload their results for a small fee, and receive their ethnic mix. We will announce this through our social channels as soon as we have an update.
What if I change my mind?
With all projects run by Living DNA if you change your mind before your data is used you can contact us and we will remove your information from the system. Data used in the projects is anonymised. Please note that this means you will not have the option to find DNA Matches, help scientific research and be part of helping to build One Family genetic tree.
We appreciate that a small number of people suggested that the information above could be made even more clear on the project website, so we are working to get these changes made by the end of the week.
Thank you for all your support in what is an incredible project that everyone can be part of,
Warmest Regards
David Nicholson & Hannah Morden
Co-Founders - Living DNA
Disclosure. I shared my raw data files from AncestryDNA, 23andMe and FTDNA with Living DNA to help them test their system prior to the launch of their test in September 2016. In return I received a free Living DNA test. All views expressed here are my own.
Wednesday, 9 August 2017
23andMe launch a new v5 chip and revise their health and trait offerings in the UK
23andMe have quietly rolled out their new v5 chip. There has not so far been any official announcement from 23andMe, though the news was confirmed yesterday by a moderator in the 23andMe Forums. We believe 23andMe are now using Illumina's new Global Screening Array chip which is already being used by Living DNA. 23andMe were one of the 12 customers who signed up to the GSA in June 2016. They also belong to the Global Screening Array Consortium.
The GSA has 640,000 markers and the ability to include up to 50,000 custom markers. Here is a description of the chip from Illumina:
The Illumina Omniexpress chip, which was previously used by all the genetic genealogy companies, is being phased out, so we are likely to see other companies moving to the GSA in due course. The OmniExpress worked well for European populations but was not so good for other populations. The GSA provides much better global coverage and should improve the results for people with non-European ancestry.
The GSA is designed for imputation. Imputation is the process of inferring the missing markers in a DNA sequence. This can be done by statistical methods because DNA is passed on in chunks, which means that markers travel together. Researchers can use imputation to reconstruct an entire genome sequence, and it provides a much more cost-efficient way of doing large-scale studies. However, I know of no scientific papers which have looked at the efficacy of imputation for cousin matching. The companies will have a significant challenge ahead in the months to come as they adapt to the new chip and test out their imputation pipelines.
Because there are so few overlapping markers between the GSA and the OmniExpress this change will also present problems for companies and third-party websites that accept autosomal DNA transfers. A choice will need to be made as to whether to do comparisons using only the overlapping markers or whether to experiment with imputation. GEDmatch is already experimenting with its new Genesis database, which can now accept GSA transfers. It will be interesting to see what solutions are found by other companies.
Along with the move to the new chip, 23andMe have also changed their product offering in the UK. There has not been any official announcement but as far as I can establish from the archived versions of their British website in the Internet Archive the changeover seems to have occurred some time towards the beginning of May. When the website was archived on 29th April 23andMe were still offering UK customers 100 health and trait reports. They offered 40+ reports for inherited conditions, 40+ drug response reports, 10+ genetic risk factor reports and 40+ trait reports. The genetic risk factor reports including reports for haemophilia and some breast cancer genes (BRCA1 and BRCA 2). A full list of the previously offered health reports can be seen here.
23andMe have slowly been moving their customers over to their new website (otherwise known as The New Experience or TNE for short). My accounts were finally transitioned on 15th June 2017. I understand that all the UK accounts were transitioned at the same time. I believe the transition process is now complete, with the Canadians appearing to be the last people to be transitioned in July.
The GSA has 640,000 markers and the ability to include up to 50,000 custom markers. Here is a description of the chip from Illumina:
The Infinium Global Screening Array-24 v1.0 BeadChip combines multi-ethnic genome-wide content, curated clinical research variants, and quality control (QC) markers for precision medicine research.
The genome-wide content was selected for high imputation accuracy at minor allele frequencies of >1% across all 26 1000 Genomes Project populations. The clinical research content includes variants with established disease associations, relevant pharmacogenomics markers, and curated exonic content based on ClinVar, NHGRI, PharmGKB, and ExAC databases. Quality control content enables sample identification and tracking for large-scale genomics and screening applications.The full technical specifications for the GSA chip can be found here on the Illumina website.
The Illumina Omniexpress chip, which was previously used by all the genetic genealogy companies, is being phased out, so we are likely to see other companies moving to the GSA in due course. The OmniExpress worked well for European populations but was not so good for other populations. The GSA provides much better global coverage and should improve the results for people with non-European ancestry.
The GSA is designed for imputation. Imputation is the process of inferring the missing markers in a DNA sequence. This can be done by statistical methods because DNA is passed on in chunks, which means that markers travel together. Researchers can use imputation to reconstruct an entire genome sequence, and it provides a much more cost-efficient way of doing large-scale studies. However, I know of no scientific papers which have looked at the efficacy of imputation for cousin matching. The companies will have a significant challenge ahead in the months to come as they adapt to the new chip and test out their imputation pipelines.
Because there are so few overlapping markers between the GSA and the OmniExpress this change will also present problems for companies and third-party websites that accept autosomal DNA transfers. A choice will need to be made as to whether to do comparisons using only the overlapping markers or whether to experiment with imputation. GEDmatch is already experimenting with its new Genesis database, which can now accept GSA transfers. It will be interesting to see what solutions are found by other companies.
Along with the move to the new chip, 23andMe have also changed their product offering in the UK. There has not been any official announcement but as far as I can establish from the archived versions of their British website in the Internet Archive the changeover seems to have occurred some time towards the beginning of May. When the website was archived on 29th April 23andMe were still offering UK customers 100 health and trait reports. They offered 40+ reports for inherited conditions, 40+ drug response reports, 10+ genetic risk factor reports and 40+ trait reports. The genetic risk factor reports including reports for haemophilia and some breast cancer genes (BRCA1 and BRCA 2). A full list of the previously offered health reports can be seen here.
 |
| The 23andMe GB home page on 29th April 2107. |
The next archived version of the 23andMe GB website dates from 6 May 2017. From that date onwards UK customers have been offered a very much reduced range of health reports. There are now 40+ carrier status reports, 3+ genetic risk reports and 25+ traits and wellness reports. The drug response reports are no longer provided at all, and the breast cancer and haemophilia reports have been dropped. A full list of the new reports offered can be found here. As far as I can establish UK customers now receive exactly the same version of the test as customers in the US, the only difference being that we do not have an option to order an ancestry-only version of the test for half the price.
 |
| The 23andMe GB home page on 9th August 2017. |
23andMe have slowly been moving their customers over to their new website (otherwise known as The New Experience or TNE for short). My accounts were finally transitioned on 15th June 2017. I understand that all the UK accounts were transitioned at the same time. I believe the transition process is now complete, with the Canadians appearing to be the last people to be transitioned in July.
With the transfer to the new website all our old health reports have been archived. There are no new health reports. Instead there are five ancestry reports, 19 trait reports and 7 "wellness" reports.
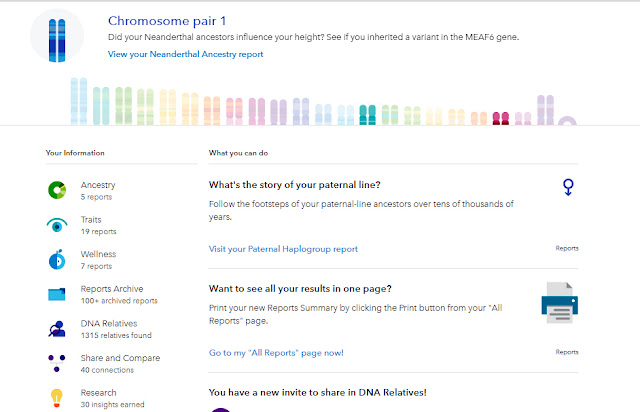 |
I was on holiday in the US when I was transitioned and I initially assumed that the lack of new health reports was because I was accessing the website from the US and not the UK. However, I later established that none of the transitioned accounts in the UK were receiving any new health reports. I wrote to 23andMe customer service to find out why this was the case and received the following message on 10th July:
Per FDA restrictions, newly authorized reports will not be provided to customers genotyped on your current chip version. The conditions covered by our newly authorized reports are addressed by reports in your Reports Archive.My guess is that 23andMe customers in the UK have been tested on the new v5 chip from May 2017 onwards when the website changed and the number of reports was reduced.
In order to receive the newly authorized reports, you will need to be genotyped again. Upgrades are unavailable at this time. We are currently working on an upgrade policy for our customers and would encourage you to wait for this policy to be finalized. The upgrade policy will allow you to be genotyped on our most up-to-date chip at a discounted rate within the next few months. More information about upgrades will be available soon.
23andMe received authorisation from the FDA in April this year to start offering genetic health risk reports for 10 diseases and conditions. No doubt they are also working towards approval for introducing other reports too. Clearly, in order to get the FDA approval, the reports will need to be validated on the new v5 chip and it's no longer worth their while trying to update and validate the old-style health reports. I can therefore understand the decision to stop offering these reports to existing customers.
I tested with 23andMe back in 2010 on the v2 chip. I've never had to pay a subscription or pay for any updates so I've had very good value for money in the last seven years. I will be upgrading to the new chip when it becomes available to existing customers and I shall look forward to receiving all the new reports as and when they become available.
Update 10th August 2017
It has been confirmed in a post by a moderator in the 23andMe Forum that 23andMe are now using the Illumina Global Screening Array. They suggest that with an upgrade to the new chip: “Results shouldn’t change significantly, however, there may be some slight differences in Ancestry Composition, DNA Relatives, and small changes in your haplogroup assignment.”
Update 10th August 2017
It has been confirmed in a post by a moderator in the 23andMe Forum that 23andMe are now using the Illumina Global Screening Array. They suggest that with an upgrade to the new chip: “Results shouldn’t change significantly, however, there may be some slight differences in Ancestry Composition, DNA Relatives, and small changes in your haplogroup assignment.”
We are told that 23andMe have updated their phasing algorithms so that “family members can still be phased, even if they are on different chips”.
It appears that the transition to the v5 chip is not yet complete. Here is a further quote from the moderator: “Our labs are currently transitioning to the v5 chip. While transitioning, there may be some overlap between v4 and v5 data being released. Future customers will be genotyped on v5.”
It appears that the transition to the v5 chip is not yet complete. Here is a further quote from the moderator: “Our labs are currently transitioning to the v5 chip. While transitioning, there may be some overlap between v4 and v5 data being released. Future customers will be genotyped on v5.”
Tuesday, 8 August 2017
MyHeritage acquires Legacy Family Tree and discounts on Legacy webinar subscriptions
It was announced last week that MyHeritage have acquired the Millennia Corporation, the company who produce Legacy Family Tree genealogy desktop software and who also run the popular Legacy Family Tree Webinars.
For further details on this acquisition you can read the statements from MyHeritage and Legacy:
- MyHeritage Acquires the Legacy Family Tree Software and Webinar Platform MyHeritage blog, 3rd August 2017
- Legacy FamilyTree has a new home with MyHeritage Legacy News, 3 August 2017
- MyHeritage acquires Legacy Family Tree: is this good for the genealogy industry? Good for genealogists? by Thomas MacEntee, Abundant Genealogy, 4 August 2017
- MyHeritage buys Millennia by Tamura Jones, Modern Software Experience, 7 August 2017.
Most of the Legacy webinars are free to view in the first week but you need to have a subscription to access the webinar archive and some premium webinars. It's also often necessary to have a subscription to access the speakers' handouts. Much of the content up until now has been aimed at US researchers but there have also been other webinars of more general interest. I've enjoyed watching a few webinars about DNA testing, particularly those from Blaine Bettinger and Diahan Southard, who always do an excellent job of explaining complicated science in easy-to-understand language. To celebrate the acquisition there is a special offer on the annual Legacy webinar subscription and it is on sale for just $24.98 (£19) until 13th August, which is 50% off the usual price. I've decided to splash out, as there quite a few webinars I would have liked to have seen that for one reason or another I didn't have time to watch when they first became available.
AncestryDNA hits the five million milestone
Ancestry have announced on their US Facebook page that they now have an astonishing five million AncestryDNA customers. Ancestry passed the four million milestone at the end of April this year and the three million milestone in January. That means they've sold two million tests in the first seven months of 2017. If sales continue at the current rate they will have tested seven million people by the end of the year. Christmas is always the peak selling period so sales could ultimately be much higher than that. How long will it take for the database to grow to 10 million or 20 million?
AncestryDNA do not give breakdowns of sales by country. Their test has been available in the US since May 2012 but only launched in the UK in January 2015. It became available in Australia and New Zealand in May 2015, Canada in June 2015 and 29 other countries in February 2016. I would estimate that about 80% of AncestryDNA's sales are now in the US. It seems likely that a considerable amount of their growth in the last year or so has come from people testing in these new markets.
Sales seem to be particularly strong in the UK, Ireland, Australia and New Zealand. AncestryDNA have been marketing their test extensively in these countries. In the UK, for example, AncestryDNA now sponsor the long-running TV programme Long Lost Family, which regularly attracts around five million viewers. The seventh series is currently being aired on ITV, and AncestryDNA are linking their promotions to the programme. The banner below now appears when you log into your Ancestry account.
The marketing is helping to bring in a new demographic to DNA testing. People are taking a DNA test out of curiosity and not as an aid to genealogical research. However, some of these new people who are testing will start to explore their matches and become interested in family history. This can only be good news for everyone.
Note
Although Ancestry's Facebook page shows that there is a sale on in the US until 15th August, the test does not appear to be on sale in any other country at present.
Further reading
Leah La Perle Larkin has published a blog post with updated figures on the size of the GEDmatch database and an updated graph showing the growth of the autosomal DNA databases. See her article Genealogical database sizes - August 2017 update.
Update
The official AncestryDNA press release can be read here.
AncestryDNA do not give breakdowns of sales by country. Their test has been available in the US since May 2012 but only launched in the UK in January 2015. It became available in Australia and New Zealand in May 2015, Canada in June 2015 and 29 other countries in February 2016. I would estimate that about 80% of AncestryDNA's sales are now in the US. It seems likely that a considerable amount of their growth in the last year or so has come from people testing in these new markets.
Sales seem to be particularly strong in the UK, Ireland, Australia and New Zealand. AncestryDNA have been marketing their test extensively in these countries. In the UK, for example, AncestryDNA now sponsor the long-running TV programme Long Lost Family, which regularly attracts around five million viewers. The seventh series is currently being aired on ITV, and AncestryDNA are linking their promotions to the programme. The banner below now appears when you log into your Ancestry account.
The marketing is helping to bring in a new demographic to DNA testing. People are taking a DNA test out of curiosity and not as an aid to genealogical research. However, some of these new people who are testing will start to explore their matches and become interested in family history. This can only be good news for everyone.
Note
Although Ancestry's Facebook page shows that there is a sale on in the US until 15th August, the test does not appear to be on sale in any other country at present.
Further reading
Leah La Perle Larkin has published a blog post with updated figures on the size of the GEDmatch database and an updated graph showing the growth of the autosomal DNA databases. See her article Genealogical database sizes - August 2017 update.
Update
The official AncestryDNA press release can be read here.
Sunday, 6 August 2017
Comparing parent and child matches at AncestryDNA
A number of genetic genealogists have done comparisons of parent and child matches at AncestryDNA to see how many of the smaller matches do not match either parent:
Ann Raymont published an article in July 2016 When is a match a false positive? She found that 35.3% of her matches did not match either parent.
Blaine Bettinger wrote a blog post on The danger of distant matches back in January this year. He found that 32% of his matches were not shared with either of his parents.
Kevin Ireland published his results in an article entitled atDNA case study: two parents and one child. He found that 18.5% of his matches did not match either parent.
Karin Lovisa Borgerson has this week published a blog post Baby versus bathwater: distant matches. She found that 17% of her matches were not shared with either parent.
I recently tested both of my parents at AncestryDNA and I thought it would be an interesting exercise to do a detailed analysis of my own matches to see how my results compare with the other studies.
I used the DNAGedcom Client to download my matches from AncestryDNA. I used the Match-O-Matic tool, which is included with the Client, to analyse my matches. See the methodology section below for details of how the analyses were done.
My matches at AncestryDNA
I tested on the AncestryDNA v1 chip in June 2012. I currently have 10,232 matches at AncestryDNA.
Using the same categories as Blaine Bettinger my matches break down like this:
Below 10 cMs the chance of not sharing a match begins to increase exponentially. With the very smallest matches sharing just 6-7 cMs only 46% matched one of my parents.
Matches that do not match my parents are either false positives, which means the matches are not real matches, or false negatives, which means that the match is not showing up in the match list of my parent for one reason or another. However, without further investigation it is not possible for me to determine whether these matches are false positives or false negatives. This can only be done by careful chromosome mapping and by testing multiple close family members.
AncestryDNA use a phased matching technique. Phasing is the process of assigning individual alleles to the maternal and paternal chromosomes. A lack of phasing results in many false matches. These are sometimes known as pseudosegments. For a discussion of the reasons for these false matches see the ISOGG Wiki article on identical by descent. Because AncestryDNA use phasing they are able to deliver matches on smaller segments than the other companies. While phasing provides more accurate matches, the process is not without its problems. One of the limitations is that we are not tested on our whole genome but rather a sampling of markers scattered across our genome. If matching were to be done on the whole genome we would no doubt find that many of our matches are not valid after all. A second problem is that the phasing algorithms are not perfect. Sometimes they break up a longer match into smaller segments. There is also a problem of what are known as phase switch errors, when the phase accidentally switches from the maternal to the paternal chromosome or vice versa.
I am fortunate that I've been able to test both my parents which allows me to do a sanity check on my matches. However, if you are not able to test your parents you will have no way of knowing which of the small segment matches are likely to be valid. It was also interesting to note that some of my matches were shared by both my parents. If I hadn't tested both my parents or if I'd only tested one of my parents I could easily have been led astray with these matches.
Matches at Family Tree DNA and 23andMe are not phased so the false match rate is going to be even higher there.
Even if these small segments are real, the odds are still stacked against the match falling within a genealogical timeframe. We know from computer simulations that over 60% of 10 cM segments are likely to trace back beyond ten generations. This does of course also mean that 40% of 10 cM matches are likely to fall within the last ten generations, but computer simulations have the advantage of working in an idealised world where every segment can be reliably attributed. In real life it is much more complicated and with the current matching algorithms there is an additional risk that these segments will not be accurately identified.
The genetic genealogy community has known for a long time the problem of using small segments in genealogical research, and my findings simply add to the existing evidence base. I already have 1773 matches at AncestryDNA that share 10 cMs or more. I can still only find genealogical connections with a handful of those matches. There is really no reason to get down in the weeds with these small segments under 10 cMs.
Update
Since publishing my blog post Alex Coles has also done an analysis of her parent and child matches at AncestryDNA. Alex and her parents all tested on the v1 chip. Alex found that 31% of her matches did not match either parent. All the non-matches were below 17 cMs apart from one intriguing outlier. Read Alex's article Imprecise science. Part 1 AncestryDNA on her Winging It blog
Related blog posts
My matches at AncestryDNA
I tested on the AncestryDNA v1 chip in June 2012. I currently have 10,232 matches at AncestryDNA.
Using the same categories as Blaine Bettinger my matches break down like this:
| Category | Number | Percentage |
| Over 50 cMs | 9 | 0.08% |
| 25 cMs or more | 39 | 0.38% |
| 20 cMs or more | 71 | 0.69% |
| 15 cMs or more | 251 | 2% |
| 10 cMs or more | 1403 | 14% |
| Fewer than 10 cMs | 8837 | 86% |
| 6-7 cMs | 4512 | 44% |
Two of my matches in the over 50 cMs category are my parents. All the other matches have tested independently at AncestryDNA.
Sharing with my
parents at AncestryDNA
My parents were tested on the AncestryDNA v2 chip in June 2017
My dad has 8350 matches at AncestryDNA.
My dad has 8350 matches at AncestryDNA.
My mum has 11285 matches at AncestryDNA.
Here are my findings after comparing my matches with my mum and dad:
- 3299 (32%) of my 10232 matches are shared with my dad
- 3276 (32%) of my 10232 matches are shared with mum
- 20 (0.2%) of my matches appear on the match lists of both my mum and my dad.
- 3671 (36%) of my matches do not appear on the match lists of either of my parents.
- Of the 3671 matches which do not match either of my parents 3559 (97%) shared a single DNA segment and 112 (3%) shared 2 segments. Ninety-six (86%) of the two segment matches shared less than 10 cMs, and 16 (14%) shared between 10 and 16 cMs.
I divided the matches into "bins" to see what the match rate was for different levels of sharing. The results are shown in the table below.
| cM bins | Total matches |
Total matching a parent |
% matching a parent |
Total matching neither parent |
% matching neither parent |
| 50 cMs + | 9 | 9 | 100% | 0 | 0% |
| 40-50 cMs | 1 | 1 | 100% | 0 | 0% |
| 30-40 cMs | 10 | 10 | 100% | 0 | 0% |
| 20-30 cMs | 51 | 51 | 100% | 0 | 0% |
| 19-20 cMs | 25 | 25 | 100% | 0 | 0% |
| 18-19 cMs | 18 | 17 | 94% | 1 | 6% |
| 17-18 cMs | 30 | 30 | 100% | 0 | 0% |
| 16-17 cMs | 46 | 45 | 98% | 1 | 2% |
| 15-16 cMs | 61 | 58 | 95% | 3 | 5% |
| 14-15 cMs | 96 | 86 | 90% | 10 | 10% |
| 13-14 cMs | 148 | 134 | 91% | 14 | 9% |
| 12-13 cMs | 180 | 161 | 89% | 19 | 11% |
| 11-12 cMs | 291 | 274 | 94% | 17 | 6% |
| 10-11 cMs | 437 | 386 | 88% | 51 | 12% |
| 9-10 cMs | 799 | 679 | 85% | 120 | 15% |
| 8-9 cMs | 1275 | 1001 | 79% | 274 | 21% |
| 7-8 cMs | 2241 | 1534 | 68% | 707 | 32% |
| 6-7 cMs | 4512 | 2058 | 46% | 2454 | 54% |
Double matches
I also took a look at the 20 matches that my parents shared with each other. My parents do not appear as matches to each other and do not have any identifiable common genealogical ancestors. I also checked to see if these shared matches appeared on my match list. Here is the breakdown:
| Match | cMs shared with Dad |
cMs shared with Mum |
Match to Debbie |
| Match 1 | 8.7576 | 7.9563 | Yes |
| Match 2 | 9.2653 | 6.984 | Yes |
| Match 3 | 9.3549 | 7.363 | No |
| Match 4 | 6.082 | 7.5295 | Yes |
| Match 5 | 6.4811 | 8.0814 | No |
| Match 6 | 9.3519 | 7.4345 | Yes |
| Match 7 | 27.0902 | 8.1206 | No |
| Match 8 | 10.009 | 14.0302 | Yes |
| Match 9 | 6.5761 | 9.6979 | Yes |
| Match 10 | 6.262 | 7.8819 | No |
| Match 11 | 6.4201 | 7.5106 | No |
| Match 12 | 7.9815 | 6.1787 | Yes |
| Match 13 | 9.2775 | 6.3962 | No |
| Match 14 | 11.4808 | 6.3302 | Yes |
| Match 15 | 7.26 | 6.1234 | Yes |
| Match 16 | 6.8033 | 7.0212 | Yes |
| Match 17 | 8.8658 | 6.2839 | Yes |
| Match 18 | 7.1988 | 6.0201 | Yes |
| Match 19 | 6.5682 | 8.9985 | Yes |
| Match 20 | 10.7345 | 14.5475 | Yes |
Discussion
Although 36% of my matches did not match either of my parents, the headline figure is not as gloomy as it might at first appear. The vast majority of these non-matches were on small segments under 10 cMs, and the lion's share of non-matches were on the very tiny segments under 7 cMs.
All matches sharing 19 cMs or more were shared with one of my parents so this can be considered my personal safe zone where matches are guaranteed to be valid.
All matches sharing 19 cMs or more were shared with one of my parents so this can be considered my personal safe zone where matches are guaranteed to be valid.
There were just four out of my 155 matches sharing between 15 and 19 cMs which did not match one of my parents. The largest non-shared match was 18.2 cMs. This means that 87.5% of my matches in this range were valid.
Below 10 cMs the chance of not sharing a match begins to increase exponentially. With the very smallest matches sharing just 6-7 cMs only 46% matched one of my parents.
Matches that do not match my parents are either false positives, which means the matches are not real matches, or false negatives, which means that the match is not showing up in the match list of my parent for one reason or another. However, without further investigation it is not possible for me to determine whether these matches are false positives or false negatives. This can only be done by careful chromosome mapping and by testing multiple close family members.
AncestryDNA use a phased matching technique. Phasing is the process of assigning individual alleles to the maternal and paternal chromosomes. A lack of phasing results in many false matches. These are sometimes known as pseudosegments. For a discussion of the reasons for these false matches see the ISOGG Wiki article on identical by descent. Because AncestryDNA use phasing they are able to deliver matches on smaller segments than the other companies. While phasing provides more accurate matches, the process is not without its problems. One of the limitations is that we are not tested on our whole genome but rather a sampling of markers scattered across our genome. If matching were to be done on the whole genome we would no doubt find that many of our matches are not valid after all. A second problem is that the phasing algorithms are not perfect. Sometimes they break up a longer match into smaller segments. There is also a problem of what are known as phase switch errors, when the phase accidentally switches from the maternal to the paternal chromosome or vice versa.
I am fortunate that I've been able to test both my parents which allows me to do a sanity check on my matches. However, if you are not able to test your parents you will have no way of knowing which of the small segment matches are likely to be valid. It was also interesting to note that some of my matches were shared by both my parents. If I hadn't tested both my parents or if I'd only tested one of my parents I could easily have been led astray with these matches.
Matches at Family Tree DNA and 23andMe are not phased so the false match rate is going to be even higher there.
Even if these small segments are real, the odds are still stacked against the match falling within a genealogical timeframe. We know from computer simulations that over 60% of 10 cM segments are likely to trace back beyond ten generations. This does of course also mean that 40% of 10 cM matches are likely to fall within the last ten generations, but computer simulations have the advantage of working in an idealised world where every segment can be reliably attributed. In real life it is much more complicated and with the current matching algorithms there is an additional risk that these segments will not be accurately identified.
The genetic genealogy community has known for a long time the problem of using small segments in genealogical research, and my findings simply add to the existing evidence base. I already have 1773 matches at AncestryDNA that share 10 cMs or more. I can still only find genealogical connections with a handful of those matches. There is really no reason to get down in the weeds with these small segments under 10 cMs.
Methodology
I am using version 1.5.1.3 of the DNAGedcom Client with a PC running Windows 7. The DNAGedcom Client is a subscription service costing $5 a month. The Match-O-Matic tool is included in the subscription. Match-O-Matic was designed for a Mac but converted to a
Windows format by Rob Warthen for use in the DNAGedcom Client. For details of
the DNAGedcom Client and Match-O-Matic see the user guide.
I downloaded my match lists into Excel spreadsheets using
the DNAGedcom client on 4th and 5th August.
I used the Match-O-Matic tool provided with the DNAGedcom client
to analyse my matches.
To see how many matches I shared with my parents I used the
report labelled Matches in common (matches in both files) [ICW] to combine the match lists for my mum and dad..
To see how many matches did not appear in either of my
matches lists I then used the report labelled Combine files (all matches without
duplicates) [ALL]. In order to get the program to work correctly I renamed the output file with the prefix m_.
I used the report labelled Matches in A that are not in
b [ANB] to extract a list of matches that were in my match list but were not in
the combined match list of my parents.
Acknowledgements
Thank you to Rob Warthen for developing the DNAGedcom
Client. Thank you to Don Worth for developing the Match-O-Matic. Thank you to
Richard Weiss for advice on using Match-O-Matic.Update
Since publishing my blog post Alex Coles has also done an analysis of her parent and child matches at AncestryDNA. Alex and her parents all tested on the v1 chip. Alex found that 31% of her matches did not match either parent. All the non-matches were below 17 cMs apart from one intriguing outlier. Read Alex's article Imprecise science. Part 1 AncestryDNA on her Winging It blog
Related blog posts
Subscribe to:
Comments (Atom)
















